Hai Duong: Hep B and BCG
Duc Du, Thinh Ong
2020-01-29 (update: 2022-08-23)
Last updated: 2022-08-23
Checks: 6 1
Knit directory: Vaccination_COVID/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has staged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210126) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d52958b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: code/.DS_Store
Unstaged changes:
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-12-1.png
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-13-1.png
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-2-1.png
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-4-1.png
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-5-1.png
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-6-1.png
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-7-1.png
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-8-1.png
Deleted: analysis/hepb_bcg_files/figure-html/unnamed-chunk-9-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-10-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-11-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-12-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-13-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-14-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-15-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-16-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-17-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-24-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-27-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-28-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-3-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-4-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-5-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-6-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-7-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-8-1.png
Deleted: analysis/measles_files/figure-html/unnamed-chunk-9-1.png
Staged changes:
New: analysis/check_haiduong.Rmd
New: analysis/check_haiduong.docx
Modified: analysis/clean.Rmd
New: analysis/hepb_bcg.Rmd
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-12-1.png
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-13-1.png
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-2-1.png
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-4-1.png
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-5-1.png
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-6-1.png
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-7-1.png
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-8-1.png
New: analysis/hepb_bcg_files/figure-html/unnamed-chunk-9-1.png
Modified: analysis/measles.Rmd
New: analysis/measles_files/figure-html/unnamed-chunk-10-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-11-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-12-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-13-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-14-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-15-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-16-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-17-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-24-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-27-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-28-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-3-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-4-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-5-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-6-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-7-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-8-1.png
New: analysis/measles_files/figure-html/unnamed-chunk-9-1.png
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
knitr::opts_chunk$set(echo = FALSE, warning = FALSE, message = FALSE, out.width = "100%", root.dir = rprojroot::find_rstudio_root_file())
library(data.table)
library(dplyr)Warning: package 'dplyr' was built under R version 4.1.2
Attaching package: 'dplyr'The following objects are masked from 'package:data.table':
between, first, lastThe following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(tidyr)Warning: package 'tidyr' was built under R version 4.1.2library(lubridate)
Attaching package: 'lubridate'The following objects are masked from 'package:data.table':
hour, isoweek, mday, minute, month, quarter, second, wday, week,
yday, yearThe following objects are masked from 'package:base':
date, intersect, setdiff, unionlibrary(ggplot2)
library(gtsummary)Warning: package 'gtsummary' was built under R version 4.1.2library(ggsci)
datap <- file.path("~", "Downloads", "updated_dataset")
bcg <- readRDS(file.path(datap, "bcg_haiduong.rds"))
bcg <- data.table(bcg)
bcg <- bcg[which(bcg$shot == 1),]
hepb <- readRDS(file.path(datap, "hepb_haiduong.rds"))
hepb <- data.table(hepb)
hepb <- hepb[which(hepb$shot == 1),]
measle_all <- readRDS(file.path(datap, "measles_haiduong.rds"))
measle_all <- data.table(measle_all)
time_step <- "month"
bcg$vacym <- floor_date(bcg$vacdate, time_step)
hepb$vacym <- floor_date(hepb$vacdate, time_step)Hep B
Number of days from Hep B vaccination to date of birth 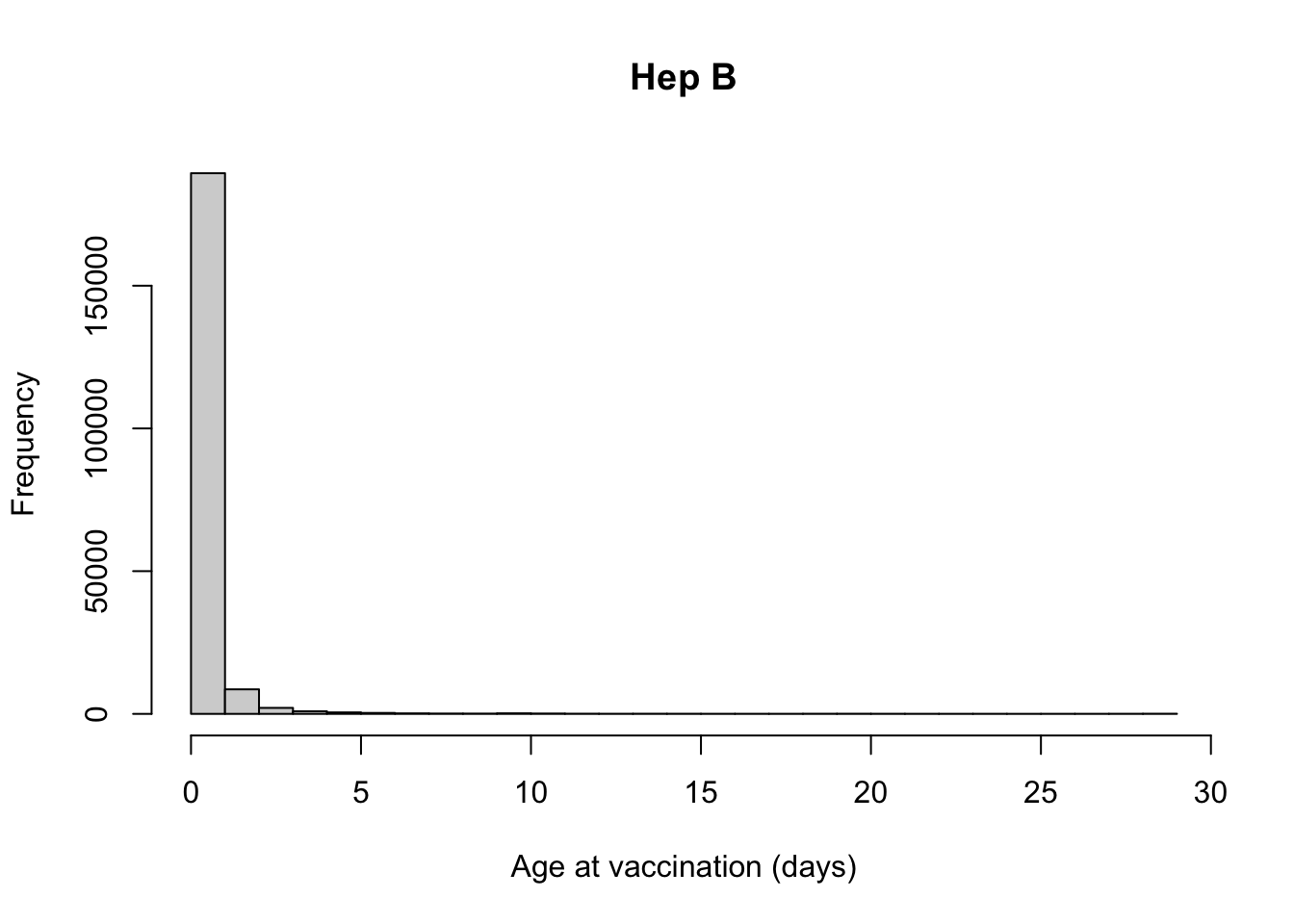
Var1 Freq
212 0 96114
213 1 93316
214 2 8599
215 3 2099
216 4 907
217 5 520
218 6 331
219 7 177
220 8 114
221 9 98
222 10 199
223 11 121
224 12 62
225 13 41
226 14 53Only keep children vaccinated within 3 days after birth (should we?)
BCG
Number of days from BCG vaccination to date of birth 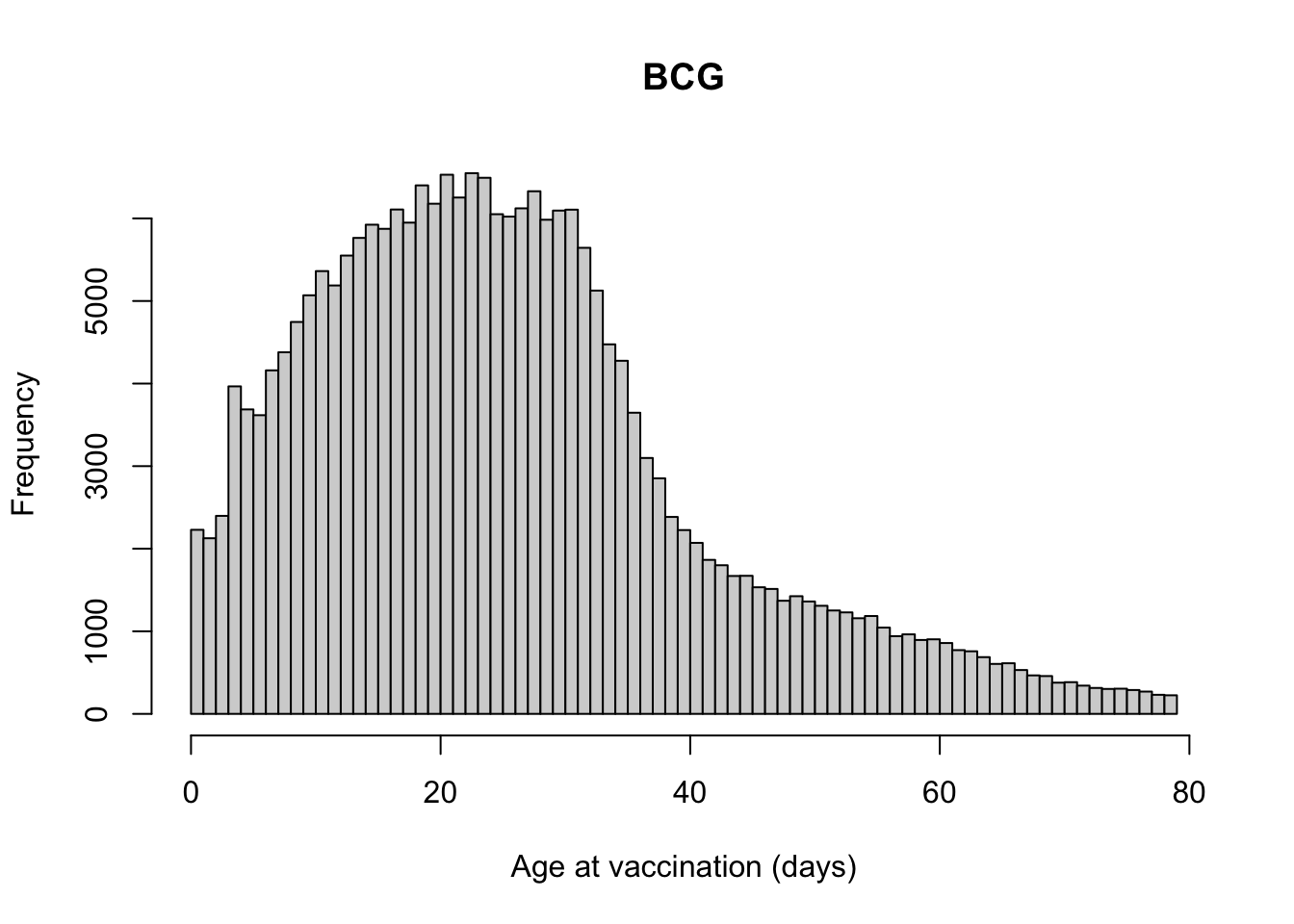
Var1 Freq
137 0 861
138 1 1368
139 2 2127
140 3 2398
141 4 3966
142 5 3687
143 6 3617
144 7 4160
145 8 4379
146 9 4746
147 10 5069
148 11 5362
149 12 5188
150 13 5550
151 14 5764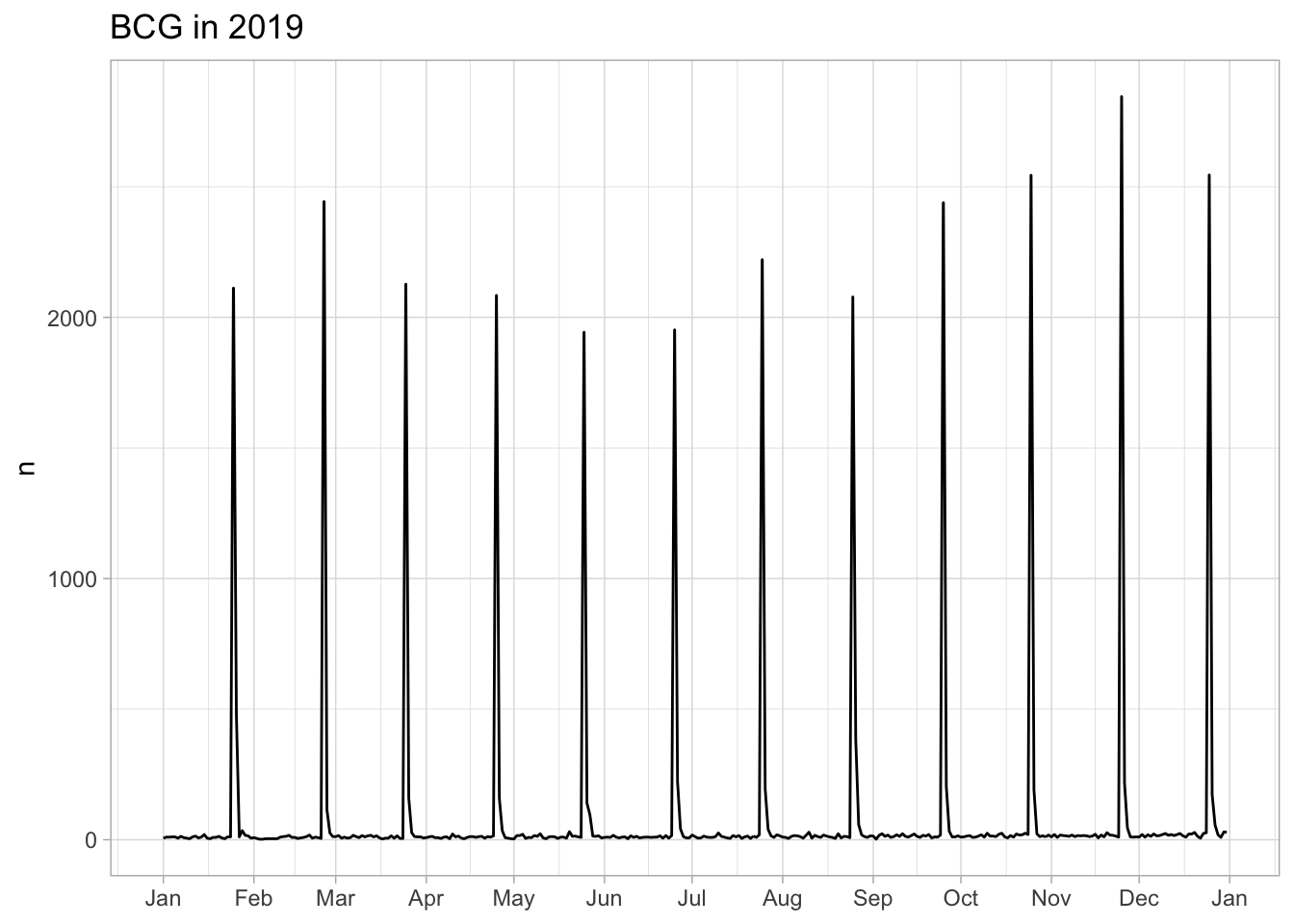
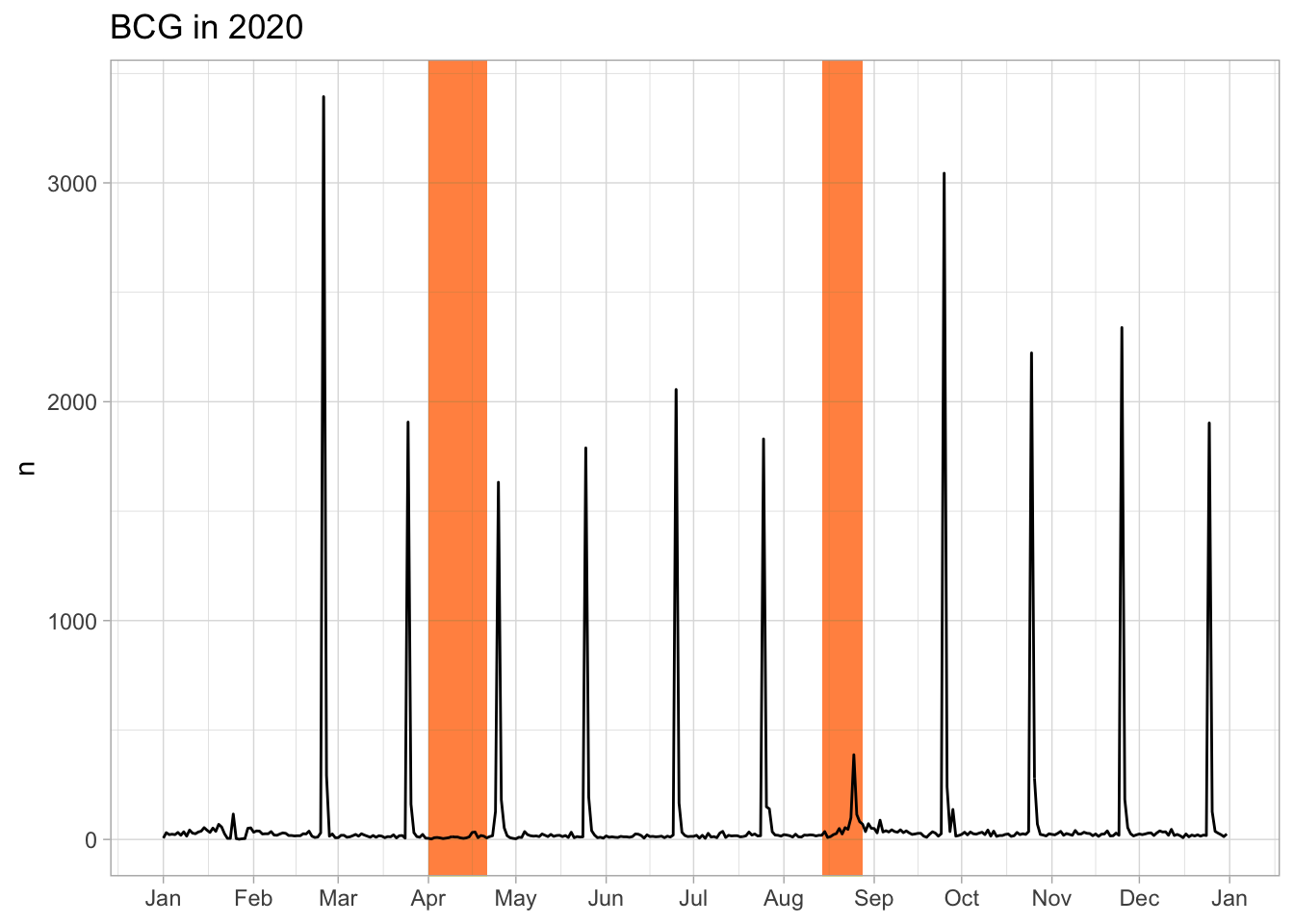
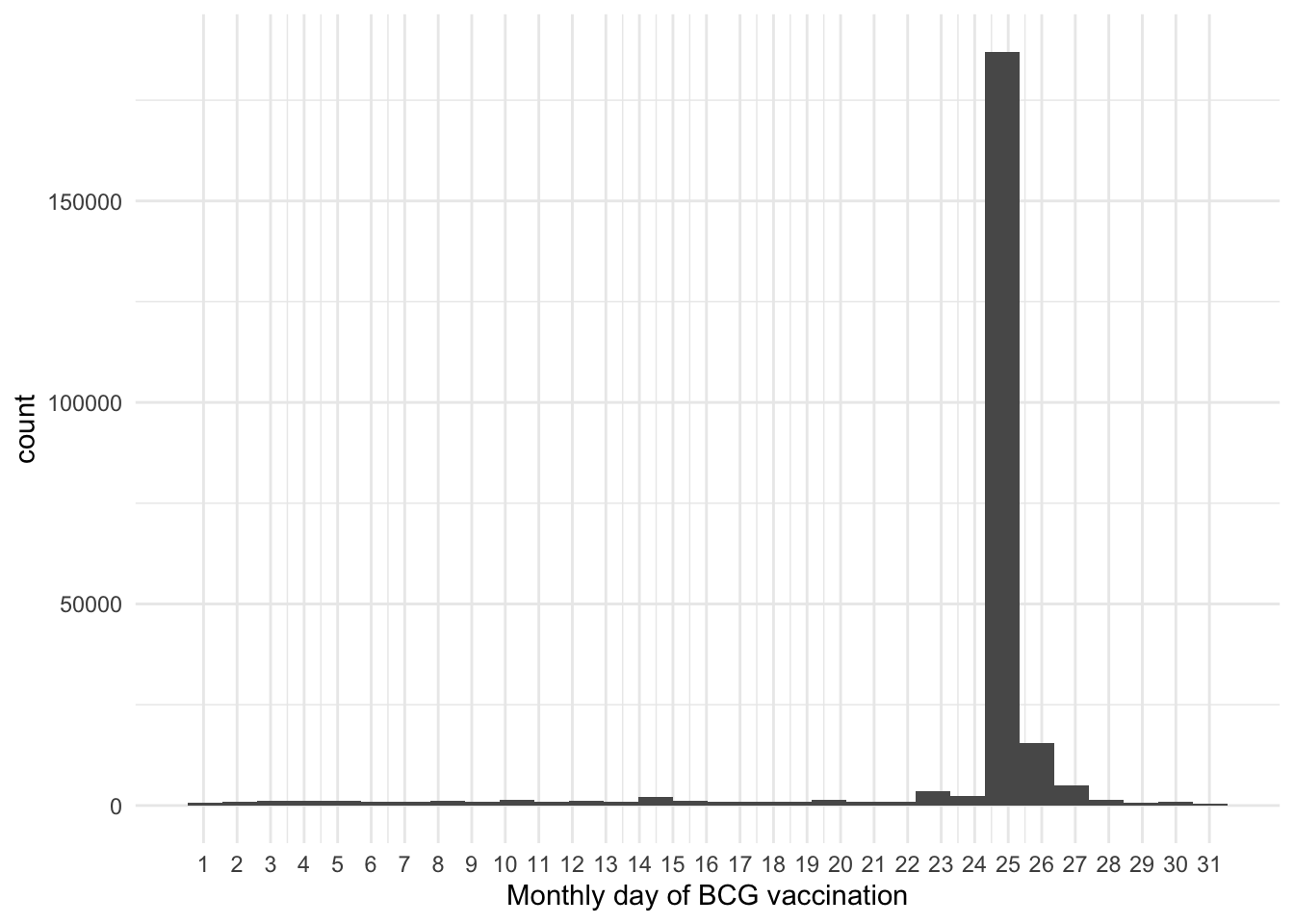
Compare Hep B and BCG
BCG in Hep B
FALSE TRUE
48813 190988 Hep B in BCG
FALSE TRUE
9140 190988 Hep B and BCG
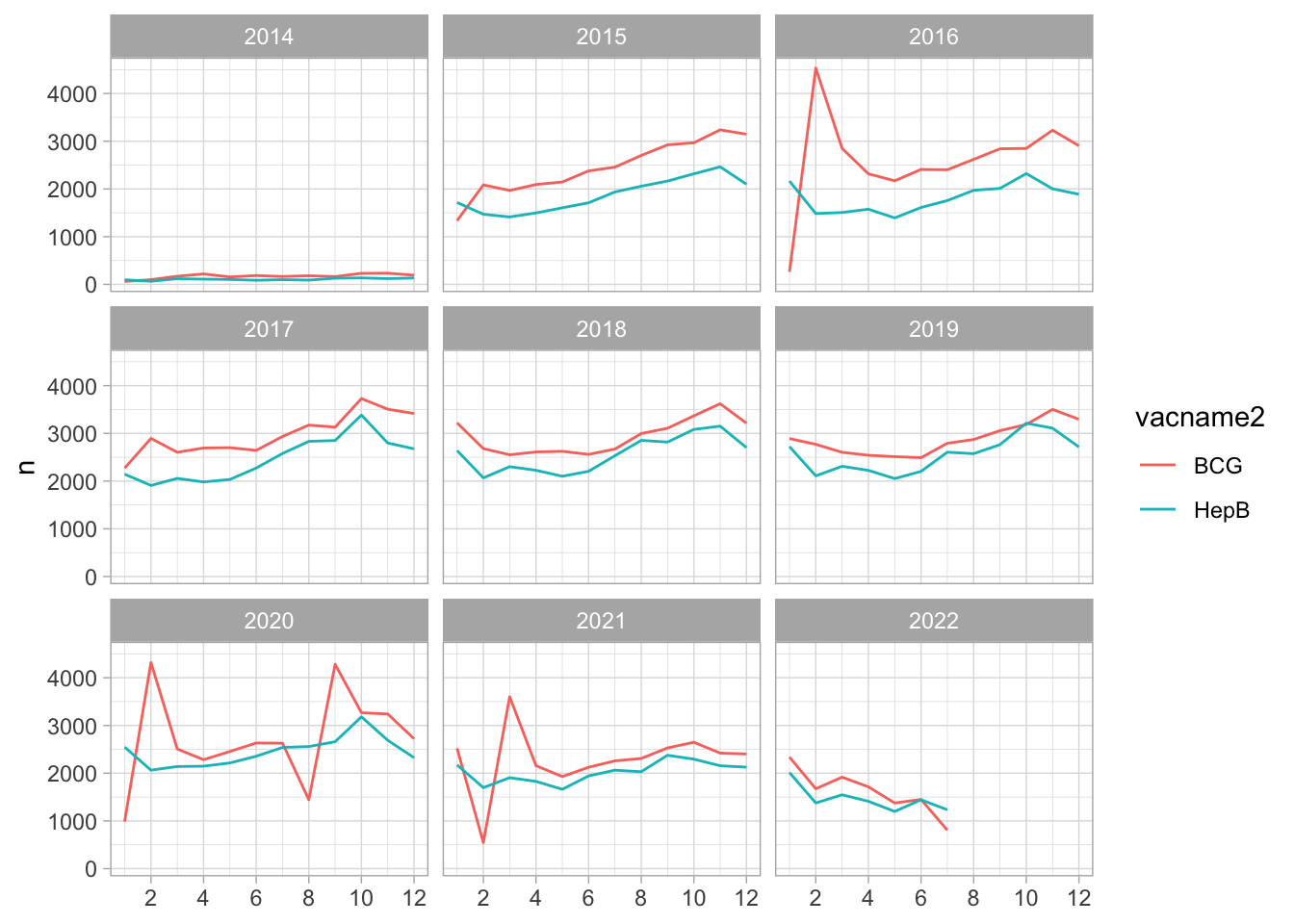
Hep B, BCG and Measles
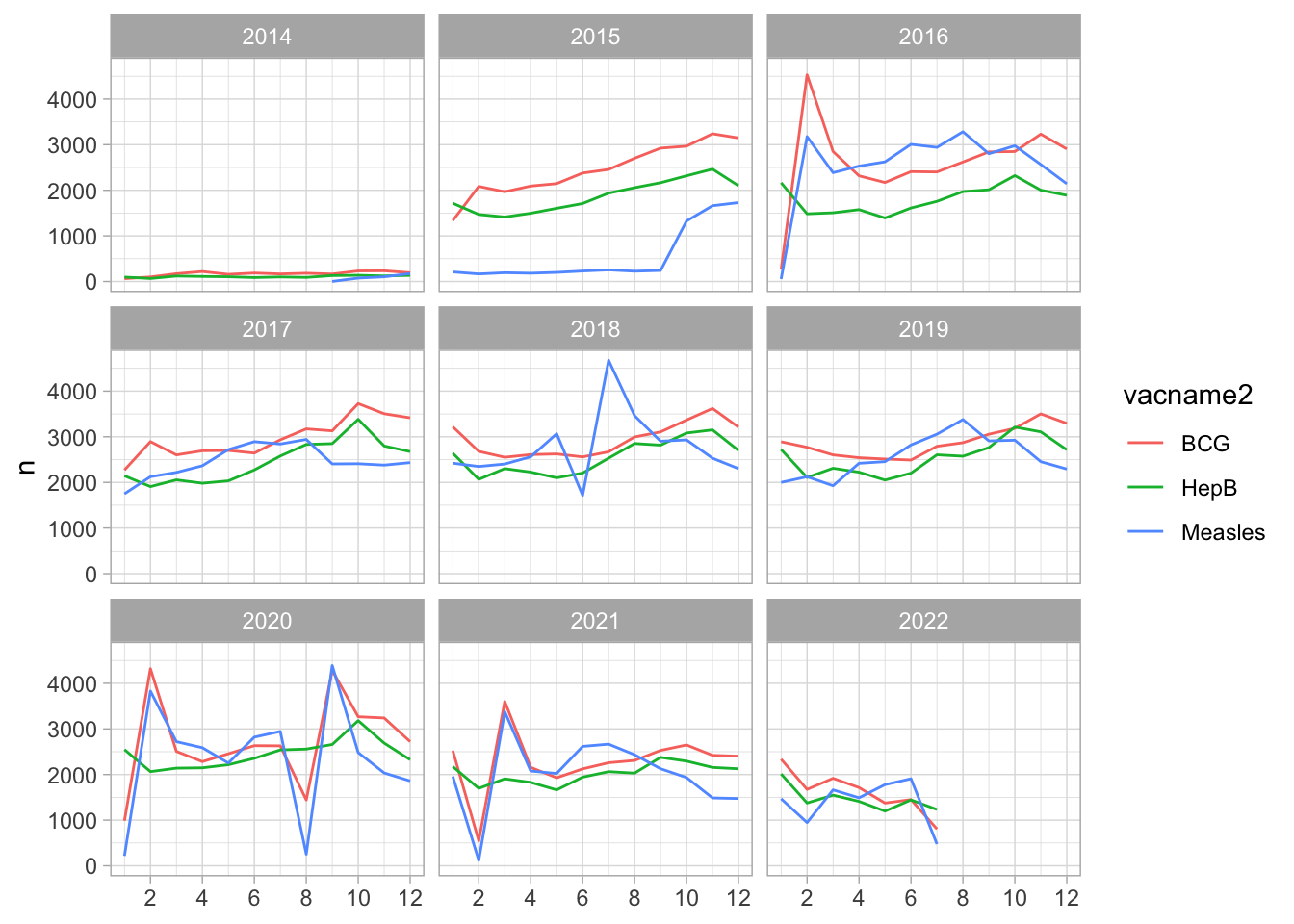
Vaccine coverage
All children born in Jan 2019 (use Hep B)
tmp1 <- hepb %>%
filter(
month(dob) == 1,
year(dob) == 2019
)
nrow(tmp1)[1] 2730Also all children born in Jan 2019 got the measles shot
tmp2 <- measle_all %>%
filter(
month(dob) == 1,
year(dob) == 2019,
vacname2 == "Measles"
) %>%
distinct(., pid, .keep_all = T)
nrow(tmp2)[1] 2729table(tmp1$pid %in% tmp2$pid)
FALSE TRUE
415 2315 2315 / 2729[1] 0.8482961These children in measles dataset get Measles shot in October 2019
tmp2 <- tmp2 %>%
filter(
vacmonth == 10,
vacyear == 2019,
)
nrow(tmp2)[1] 1360So the vaccine coverage
nrow(tmp2) / nrow(tmp1)[1] 0.4981685Cumulative coverage of children born in Jan - Feb 2019
tmp1 <- hepb %>%
filter(
month(dob) %in% c(1, 2),
year(dob) == 2019
)
tmp2 <- measle_all %>%
filter(
month(dob) %in% c(1, 2),
year(dob) == 2019,
vacname2 == "Measles"
) %>%
distinct(., pid, .keep_all = T)
nrow(tmp1)[1] 4847nrow(tmp2)[1] 4773tmp2 <- tmp2 %>%
filter(
vacmonth %in% c(10, 11),
vacyear == 2019,
)
nrow(tmp2) / nrow(tmp1)[1] 0.6769136Cumulative coverage of children born in Jan - Feb - March 2019
tmp1 <- hepb %>%
filter(
month(dob) %in% c(1, 2, 3),
year(dob) == 2019
)
tmp2 <- measle_all %>%
filter(
month(dob) %in% c(1, 2, 3),
year(dob) == 2019,
vacmonth %in% c(10, 11, 12),
vacyear == 2019,
vacname2 == "Measles"
) %>%
distinct(., pid, .keep_all = T)
nrow(tmp2) / nrow(tmp1)[1] 0.7470621
R version 4.1.1 (2021-08-10)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggsci_2.9 gtsummary_1.6.1 ggplot2_3.3.5 lubridate_1.8.0
[5] tidyr_1.2.0 dplyr_1.0.9 data.table_1.14.2
loaded via a namespace (and not attached):
[1] tidyselect_1.1.2 xfun_0.32 bslib_0.4.0
[4] purrr_0.3.4 colorspace_2.0-3 vctrs_0.4.1
[7] generics_0.1.3 htmltools_0.5.3 yaml_2.3.5
[10] utf8_1.2.2 rlang_1.0.4 jquerylib_0.1.4
[13] later_1.3.0 pillar_1.8.1 glue_1.6.2
[16] withr_2.5.0 DBI_1.1.3 plyr_1.8.7
[19] lifecycle_1.0.1 stringr_1.4.1 munsell_0.5.0
[22] gtable_0.3.0 workflowr_1.7.0 evaluate_0.16
[25] labeling_0.4.2 knitr_1.39 fastmap_1.1.0
[28] httpuv_1.6.5 fansi_1.0.3 highr_0.9
[31] Rcpp_1.0.9 promises_1.2.0.1 scales_1.2.1
[34] cachem_1.0.6 jsonlite_1.8.0 farver_2.1.0
[37] fs_1.5.2 digest_0.6.29 stringi_1.7.8
[40] rprojroot_2.0.3 grid_4.1.1 cli_3.3.0
[43] tools_4.1.1 magrittr_2.0.3 sass_0.4.2
[46] tibble_3.1.8 crayon_1.5.1 pkgconfig_2.0.3
[49] broom.helpers_1.8.0 assertthat_0.2.1 gt_0.6.0
[52] rmarkdown_2.15 rstudioapi_0.13 R6_2.5.1
[55] git2r_0.30.1 compiler_4.1.1